The general aim of my research is to achieve scientific breakthroughs and develop innovative methods, algorithms and technologies using interdisciplinary research in applied mathematics, algorithms, computational theory and biochemistry with applications to protein science, data science and robotics.
As an applied mathematician and computational biologist, my research spans several areas of bioinformatics and discrete applied mathematics, specializing in algorithm development to understand how proteins function. The techniques I use and develop are at the confluence of structural-mathematical rigidity theory, discrete applied mathematics (graph theory, discrete and computational geometry, combinatorial algorithms, matroid theory), and various tools and methods in computational biology, bioinformatics and machine learning.
I am very much attracted to research problems in which I can solve fundamental biological problems, particularly relating to protein structures and dynamics, while pushing the state of the art in mathematical theory and algorithms. I develop and apply computational and mathematical methods which are combined with experiments to understand how proteins perform their function. These mathematical techniques and models also have applications in robotics and mechanical linkages research which is another theme of my research. My research involves a large number of international collaborations with computational biologists, experimentalists, mathematicians and robotics researchers (please see below). I specialize in the development of custom-built algorithms (often inspired from mathematical rigidity theory) and computational/bioinformatics methods to unravel dynamic character of protein structures (or robotic structures). If we can understand protein dynamics and its flexibility at atomistic level, we can ultimately decipher their biological functions. While protein dynamics and flexibility can be studied with experimental techniques and traditional molecular dynamics simulations, they are often costly, provide limited information and are computationally impractical. My algorithms and methods address critical computational bottlenecks which can rapidly and accurately elucidate the complex interplay between structural rigidity, flexibility, dynamics and function of proteins, which has led to several breakthrough in structural biology.
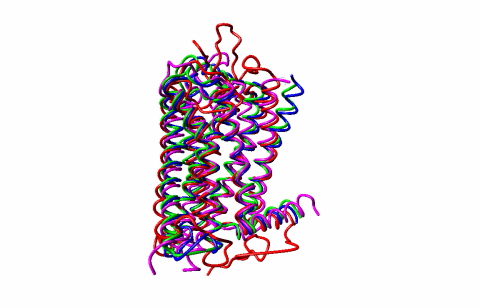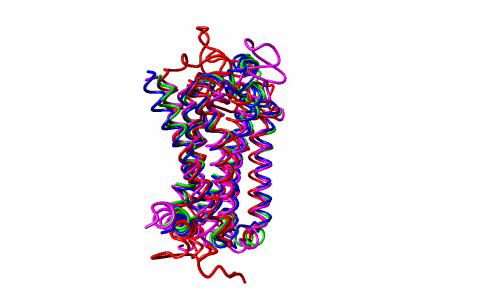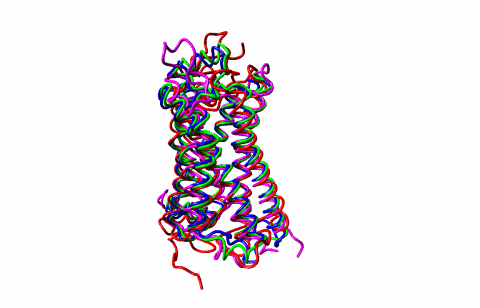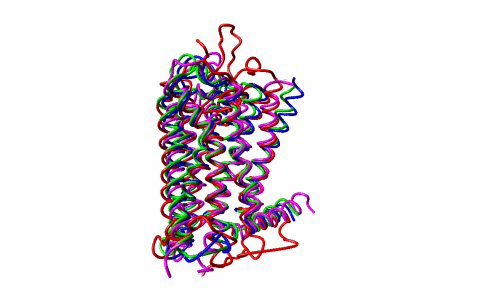
I showed how enzymes utilize allostery to perform catalysis and the role of allostery in activation of largest class of human receptors and drug targets G-Protein Coupled Receptors (GPCRs). These studies were published in Science, Cell, Nature Communications and J. Am. Chem Soc. I have also developed and applied computational tools for fast prediction of protein flexibility and dynamics which have provided new insights into how antibodies interact with antigens and how chemical defects in intrinsically disordered proteins such as TAU (key protein in Alzheimer’s) affect their shapes and function. With a group at Sheffield University, I have developed a first workable method for validation of NMR protein structures (published in Nature Communications 2020), which is a major advance in protein structure biology.
Some of my current research projects:
1. Deciphering a general mechanism of allostery in proteins (multiple collaborations in Japan, Europe, USA and Canada):
Allostery, which has been coined the “second secret of life”, refers to regulation of protein function by transfer of signals between distant sites on protein structure. Modeling allostery is a difficult problem and most questions surrounding allostery are largely unresolved. A key puzzle is to describe the general physical allosteric mechanism of distant coupled conformational change and to detect regions (residues) in the protein that are the most critical for transmission of information. To address this, I have developed first rigidity mechanistic model and description of allosteric transmission during my PhD studies known as Rigidity Transmission Allostery (RTA) algorithm. We continue to investigate and develop this approach in combination with other computational methods and experimental techniques to better understand allostery.
2. Allostery and activations of G Protein Coupled Receptors (GPCRs) ( See Press Release on activation of GPCRs.)
( See Press Release on role of calcium and magnesium in activation of GPCRs.)
3. Fast computational predictions of protein flexibility, ensembles and its dynamics
4. Protein structure validations and refinement
( See Press Release on development of method for validation of NMR protein structures.)
5. Intrinsically disordered proteins: conformational ensemble and dynamics
6. SARS-CoV-2 protein dynamics analysis
7. Geometry-based Molecular Dynamics simulations and reinforcement learning
8. Evolution of Antibody flexibility and mechanism of affinity maturation (collaboration with Jeffrey Gray at John Hopkins University and University of Tokyo)
9. Algorithms and analysis for design of mechanical linkages, robots and metamaterials (collaboration with Andreas Muller at Johannes Kepler University)
10. Autocatalytic sets and origin of life
11. Impact of symmetry on molecular flexibility and protein motions (continuation of this work).
For more information on mathematical rigidity theory see here.